Journal of Creation 24(2):13–15, August 2010
Browse our latest digital issue Subscribe
Cell systems—what’s really under the hood continues to drop jaws
Two 2009 papers summarized recent discoveries of utterly unforeseen intricacy, adaptability, robustness and precision in regulating gene expression, even in “simple” cells.
Gene expression in eukaryotic cells
I conservatively counted 24 recently discovered mechanisms that help regulate gene expression in eukaryotic cells, as reviewed by Moore and Proudfoot.1 Here are just a few of them.
Chromatin is not loosely wadded DNA inside cellular nuclei. Instead, it is very precisely organized, with specific portions dynamically looped outward. Each loop is associated with a separate nuclear pore, and can retract to a storage position when appropriate. Robust and efficient machinery ensures that the correct portions of chromatin are unspooled from nearer the center of the nucleus to an appropriate nuclear pore. Each pore is extremely active, with a host of interacting regulatory RNA’s, proteins, and ribonucleoproteins.2 These send and receive communications from and toward the farthest ends of the RNA and protein manufacturing processes.
RNA Polymerase does not typically transcribe DNA in fluid space, but is attached to a cadre of proteins associated with each nuclear pore. This way, the rapidly emerging RNA transcript is already proximal to the pore, through which much of it will exit to the cytoplasm. Further, cell biologists have determined that the first copy of a transcript is like a practice run. This first, rough draft RNA transcript either serves as a quality control run, so that its integrity is ensured prior to full manufacture and export from the nucleus, as a primer for the total set of transcript processing machinery to be properly set, as a chemical communicator providing information to downstream processes, or all three.
Warming up for transcription
In addition, extracellular messages are transferred from the cell membrane to the nuclear pore sites via biochemical cascades, and these influence whether or not a gene region will switch from being transcribed into these rough ‘abortive transcripts’, or into full-length, properly marked and exported transcripts. It appears that transcription machinery is constantly transcribing in an ‘idle’ mode, but when the correct switches are tripped, the machinery fully engages. In full production mode, RNA transcripts often become marked for translation to proteins. Some of the switching messengers are proteins that are temporarily restrained by other proteins, which in turn can release them upon detection of certain cell signals carried by yet more precisely interacting biochemicals. For example, even sugar moieties riding on proteins have been found to act as a “safety switch that regulates the microswitches” which fine tune protein expression during cell division.3
Full-on eukaryotic transcription runs super-fast
When all systems are ‘go’, transcription proceeds with “fully processive elongation of the full body of the gene.”1 Inside the nucleus, the relevant DNA is pulled, like a loop of magnetic tape, across a nuclear pore. Some of the proteins involved in this action are named Set1PAF, Spt6, FACT, Chd1, along with other histone proteins. This way, the emerging transcript is under the constant watchful attention of a wide array of sensory, quality control, marking, and transporting machinery, all kept near the pore by precise chemical interactions specified by exactly arranged biomolecular sizes, shapes, charges, and polarities.
It was known that transcripts in eukaryotic cells undergo cut-and-pasting as well as splicing. It is now known that this occurs simultaneously with manufacture, and requires a separate host of proteins. However, those pre-mRNA splicing proteins directly interact with the RNA polymerase assemblage, which all works together to react to ‘pause-sites’ in the gene it is transcribing. RNA polymerase acts like a ‘molecular juggernaut’,1 streaming RNA’s out as though through a jet engine. It must be slowed down in order for cutting and splicing machinery to have opportunity to insert. Since not all DNA pause sites become RNA cut sites, and since the alternative combinations of cut and spliced mRNA transcripts can specify a wide variety of regulatory or catalytic RNA’s and proteins from just one ‘gene’,4 it is apparent that somehow precise communication occurs to discern which pause sites will result in cuts.
In yeast, a model eukaryote, the THO/TREX protein complex serves three roles: one in transcription, one in “transcript-dependent recombination”, and one in mRNA export.1 And it does these while in constant communication with machine parts that are involved in transcript initiation as well as parts involved in slowing and stopping transcription. It is therefore one of many proteins and protein complexes that are being discovered with multiple functions—a clear sign of elegant engineering.
Process flow management in translation
The emerging RNA transcript then gets labeled with specific protein markers. The markers had already been gathered to the nuclear pore site, and are presented to the nascent transcript just inside the nucleus. The immediacy of labeling thus is vital. It guards against the dangers of having naked RNAs in the nucleus, as described below. The markers, too, serve multiple purposes. The more splices in the transcript, the more markers are attached, and this eventually causes more efficient translation because a transcript thus bedecked is more likely to have some surface exposed to cytoplasmic proteins vital to translation. The markers also signal watchdog nuclear pore proteins to expedite the transcript’s export.
These same watchdog proteins also serve to prevent naked transcripts from re-entering the nucleus. This is vital, for bits of RNA naturally anneal to unzipped DNA. If this happened, it would quickly create havoc in the nucleus by both generating mutations and gumming up the many nuclear processes that depend on accurate DNA recognition, clamping, spooling, unwinding, and other processes.
After export, the cytoplasmic machinery links each transcript to other machines. Some of these shepherd the transcript toward a ribosome. Each time a transcript has been thus shepherded, some of its markers are removed, with most being lost after its first round of translation. Eventually the transcript becomes naked and difficult for translational machinery to detect, and subject to degradation. In this way, the freshest and highest quality transcripts are by far most translated by the ribosome.
Eukaryotic gene expression is astonishing
Effective quality control mechanisms constantly cull corrupt transcripts. For example, if a transcript did not have the correct signal sequence attached when it was first formed, due to gene mutation or an error in processing, the compromised molecule would have been recognized immediately at the nuclear pore, and degraded by RNase enzymes. This ensures that downstream processes are not gummed up with useless transcripts. Quality control is critical to forming the correct products in the needed amounts, and at appropriate paces.
Other systems produce a stockpile of quality transcripts in strategic pockets within the cytoplasm. This way, there can be “a tightly controlled burst of the desired [protein] product.”1
There is no indication that the discovery pace of more mind-bogglingly brilliant cell processes will slow down anytime soon. If none of the above made sense, then let the reader be edified by the glowing research summary:
“At every point along the way, multifunctional proteins and [ribonucleoprotein] complexes facilitate communication between upstream and downstream steps, providing both feedforward and feedback information essential for proper coordination of what can only be described as an intricate and astonishing web of regulation.”1
The ‘simple’ Mycoplasma
Mycoplasma pneumoniae bacteria, long considered the simplest prokaryote, can no longer be described thus. It is a parasitic bacterium that (M. pneumonia causes walking pneumonia) has a reduced genome size. It relies on its host for certain nutrients that its ancestors apparently were able to manufacture. Thus, it has undergone significant genomic decay.
How Mycoplasma bacteria really work
The authors of a paper in Science endeavored to investigate “how a cell actually accomplishes” necessary processes using the most basic subject of study.5 But they ran into a juggernaut of layered information-rich complexity that inspired their assessment:
“Together, these findings suggest the presence of a highly structured, multifaceted regulatory machinery, which is unexpected because bacteria with small genomes contain relatively few transcription factors … revealing that there is no such a thing as a ‘simple’ bacterium.”5
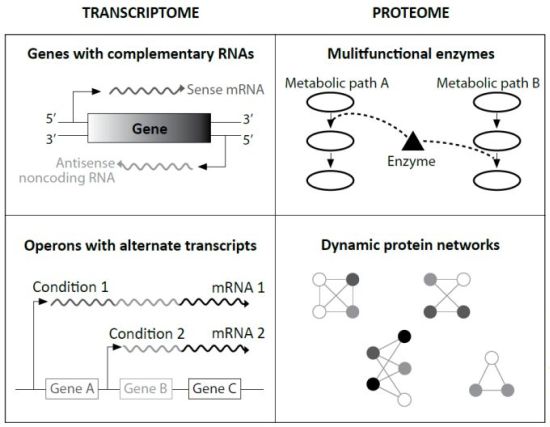
Specifically, evolutionists Ochman and Raghavan cited research that found in many cases the ‘sense’ strand of protein-coding genes is transcribed, the complementary or ‘anti-sense’ strand is also transcribed. The resulting ‘sense mRNA’ is eventually translated to protein, and the resulting ‘antisense mRNA’ binds to the sense mRNA to make a double stranded RNA. This slows its path toward translation, and is thus an important speed regulator. This was previously only known to occur in eukaryotes.
Mycoplasma cells have eukaryotic complexity
In other experiments, different environmental growth conditions caused different lengths and segments of genomic DNA to become transcribed. This implies a suite of chemical communication cascades from the cell wall inward, as well as the ability to make alternate products from one gene. This, too, was a surprise, only known in eukaryotes.
Like eukaryotic cells, these ‘simplest’ among prokaryotes have multifunctional proteins which can be used in different metabolic pathways as backup machines. Other data strongly suggests that newly manufactured proteins can be altered by other cellular machinery. Termed ‘post-translational modification’, this was taught dogmatically in my 1998 graduate biochemistry courses as exclusive to eukaryotes.
Also shocking was the discovery that over 90% of Mycoplasma proteins are involved in protein complexes, again like eukaryotes. Another genome-wide survey found indirect evidence of tight gene expression regulation, but nobody yet knows the mechanism for it.
They finally argue that because Mycoplasma is still alive even after such reduction in quality and quantity of its genome, it must have “an underlying eukaryote-like cellular organization replete with intricate regulatory networks and innovative pathways.”5
Where did Mycoplasma get all this in the first place?
These authors then bravely ask, “How did these remarkable layers of gene regulation and the highly promiscuous [multifunctional] behavior of proteins in M. pneumoniae arise?”4 But they instead explain that:
“ … the reduced efficacy of selection that operates on the genomes of host-dependent bacteria … reductions in long-term effective population size [from the bottleneck that occurred when the bacteria first became host-dependent, and] the accumulation and fixation of deleterious mutations in seemingly beneficial genes due to genetic drift, [together cause a] reducing genome size.”5
If selection, bottlenecks, and mutations only reduced the genome, then these processes are no help at all. What in nature expanded the genome with ingeniously useful data that the remarkably robust yet genomically truncated Mycoplasma retains plenty of?
Conclusion
At every level, scientists have uncovered more information. That information takes the form of three-dimensional shapes, electronic and charge configurations, as well as raw coding sequence information. Communication pathways, routines and subroutines, prioritizing, quality control, and process regulation plans are all stunningly effective and strikingly small.
More in-depth knowledge of these fantastically complicated cell features demands greater faith from naturalists in the belief that laws of chemistry built cells. The more informational structures that are found, the greater the gap between the organization in living system parts and the disorganization found in nonliving chemicals.
A reminder of some inferences about information would seem appropriate here. First, wherever precise regulation of processes due to expertly engineered machines and codes are seen coming into existence, they always comes from persons. Stated negatively, these machines and codes are never observed to originate from natural laws. Therefore, it is most parsimonious to infer that wherever similar machines, processes, and codes are found, they, too, were not derived by nature, but instead by a person or persons.
Second,
“ … like spoken languages, biological language is irreducibly complex and yet without physical substance. It comes complete with symbols, meanings for those symbols, and a grammatical structure for their interpretation. Remove any one of these three fundamental features, and the informational system is lost. Physics has nothing to do with symbols or grammar, and therefore nothing to do with the origin of life, which cannot exist without its coded information.”6
If run-of-the-mill information always comes from a mind, then this cellular information, being extraordinary, came from a mastermind.
References
- Moore, M.J. and Proudfoot, N.J., Pre-mRNA processing reaches back to transcription and ahead to translation, Cell 136:688–700, 2009. Return to text.
- Ribonucleoproteins are being found to carry out many essential functions. These are precisely arranged molecular machines comprised of partly protein, and partly RNA. The ribosome is comprised of these, but the recently discovered ribonucleoproteins are much smaller than ribosomes. Return to text.
- Sweet!―Sugar plays key role in cell division, Johns Hopkins Medicine press release, 5 February 2010, reporting research in: Wang, Z. et al., Extensive crosstalk between o-glycnacylation and phosphorylation regulates cytokinesis, Science Signaling 3(104):ra2, 2010; www.hopkinsmedicine.org/Press_releases/2010/02_05a_10.html. Return to text.
- Since the 2007 ENCODE consortium discovered that most DNA is transcribed, they recommended discontinuing the use of the word ‘gene’, which implies that only certain segments of DNA are transcribed and translated. See: The ENCODE Project Consortium, Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project, Nature 447:799–816, 2007. Return to text.
- Ochman, H. and Raghavan, R., Excavating the functional landscape of bacterial cells, Science 326:1200–1201, 2009. Return to text.
- Thomas, B., Dawkins’ latest book, The Greatest Lie on Earth, ICR News, icr.org, posted on 23 September 2009; accessed 15 December 2009. Return to text.




Readers’ comments
Comments are automatically closed 14 days after publication.