Journal of Creation 34(2):39–45, August 2020
Browse our latest digital issue Subscribe
Twelve rounds, twelve knockouts
A review of The Stairway to Life: An origin-of-life reality check by Change Laura Tan and Rob Stadler
Evorevo Books, 2020

Those who believe evolutionary theories are scientifically implausible often argue in an unsystematic manner. Frequently a ‘one-punch knockout’ strategy is used, jumping from topic to topic looking for just the right line of reasoning to avoid effort. Therefore, Dr Tan and Dr Stadler offer a structured approach to the conflict, by breaking down the naturalistic approach to the origin of life into 12 discrete stages. The starting point for each presupposes the preceding ones are fully in place. Instead of providing a single-punch knock-out argument, their careful reasonings for each stage provide an arsenal of deadly martial arts techniques.
The book is quite extraordinary. It covers so much ground in such a short volume. The authors meticulously researched the facts, documenting their most important sources in 227 references, then present the essentials in a very concise and easily understood manner. Much of the content is explained so that a high school student could grasp the key message, but the world’s top scientists will also find much source of inspiration from examples they may wish to pursue in detail.
To illustrate how a great explanation clarified an otherwise challenging topic, a merry-go-round was used as an analogy for how the ATP synthase complex works (p. 157). Each child enters and holds on to an empty seat, then contributes angular momentum when pushing off. Suddenly, everything made sense. This is quite a feat, considering that the simplest known ATP synthase consists of at least 20 interconnected proteins.
Abiogenesis research is intelligently performed
Before introducing the 12 stages natural processes would need to resolve, Tan and Stadler discuss in detail a specific example of how a cell was allegedly manufactured in a laboratory. This helps to understand some key principles, such as the need for careful planning to ensure desired intermediate outcomes, based on pre-known goals. Deep expertise is necessary coupled with chemical ‘tricks’ which are not found in nature. Most significantly, if the work resembles real cells, at some point biochemicals deriving directly from cells are needed or must be manufactured with the help of cellular materials.
The example involves a 15-year project by the J. Craig Venter Institute, using a crack team of approximately 20 scientists headed by Nobel Laureate Hamilton Smith (chapters 1–5). The hype which accompanied the publication in 2010 of what they dubbed Synthia overlooked how nothing of relevance to support evolution was accomplished. In a nutshell, the DNA sequence of a living bacterium was duplicated synthetically, and then transferred into a living cell. This would be comparable to copying a program from one computer to a different one, and then giving the impression the entire system, including hardware and operating system, could have originated on its own.
How the DNA sequence was manufactured could be too technical for some readers, who may wish to skip to the next section. Others may benefit by reflecting on how origin-of-life researchers are forced to use intelligent strategies to overcome fundamental chemical realities which otherwise produce the wrong outcomes.
No serious researcher has attempted to design a collection of novel genes (and how to regulate them) in order to create an entirely new life-form. In fact, it is common knowledge that many indispensable genes exist, in even the simplest organisms, with currently unknown functions. Therefore, the target DNA sequence matched that of an already living organism Mycoplasma mycoides (‘Myco’) almost perfectly (p. 31).
What did the team do? The target genome was split into 1,078 consecutive segments called ‘cassettes’, each containing 1,080 base pairs (p. 33). Each cassette was divided into 15 to 20 fragments of DNA, called oligonucleotides (‘oligos’). To produce each oligo, the correct sequence of deoxynucleotides (A=adenine, C=cytosine, G=guanine or T=thymine) had to be linked together. Without the cellular molecular machines which perform this linking task, Blue Heron, a company specialized in gene synthesis, had to apply some very sophisticated chemical tricks, as we will now see.
One difficulty is that the nucleotides possess several functional groups which can react together in alternative ways, all but one leading to the wrong product. To permit only the correct one to react, the others were rendered inert through chemical ‘blocking groups’. In addition, the -OH group which needed to react was activated by a special chemical substitution. These modified, fragile monomers were stored in the absence of water, air, and any impurities. Nothing like this will happen on its own in free nature.
One might be deceived into thinking that the work described so far at least involved chemicals obtained naturally, but often what is hidden under the rug is of greatest significance. It turns out that the four bonds located on some ‘chiral’ carbons of nucleotides can be arranged spatially in two manners, producing two isomers per chiral centre. These isomer pairs are mirror images of each other but are not superimposable. There are three such ‘chiral centres’ in the nucleotides of DNA, leading to eight distinct isomers. Only one of these can be used to produce DNA and it must be 100% pure. The only way to obtain the correct single isomer requires using special enzymes found in living organisms (p. 36).
The discussion so far identifies important principles. Chemists are very clever people who know how to break down problems and manipulate conditions to attain intermediate goals, step by step. None of these learned skills nor techniques, however, have any relevance to naturalism, nor demonstrate godless evolution could work.
I will only highlight the next steps used by Venter’s team as they continued to synthesize Synthia’s DNA. Living E. coli with its sophisticated polymerase molecular machine was used to produce the cassettes, yeast cells were used to combine groups of 10 cassettes, and then these latter intermediates were transferred back into E. coli.
Linking all the nucleotides using only thermal energy can only produce DNA chains a tiny fraction of a percent the necessary length, and without the accuracy necessary. Relying on complex biological molecular machines for this task illustrates once again the irrelevance of this otherwise interesting work to support evolution.
The resulting DNA sequence was still not precise enough to work due to an accidental overlooked single nucleotide deletion (p. 53) which had to be corrected. Even that was not enough, as the team discovered that the synthesized DNA had to then receive a series of methyl group attachments. How could they know what would work? Evolutionary trial-and-error, perhaps? Certainly not.
Since the DNA needed to function in an organism very similar to Myco, called Capri, they carefully mimicked the methylation patterns used by both, and transferred the modified DNA into Capri. Both patterns worked. The ‘surrogate mother’ already had all the necessary proteins together with assembled, working molecular machines in the right concentrations and locations, able to interact with the new DNA as expected.1 The last step was to set up an artificial strategy of reproductive competition to remove the original Capri DNA from Synthia.
The Synthia example, which combined world-class top-down scientific design thinking plus all the key components from pre-existing living cells, prepares the reader’s thinking for why naturalism fails badly in accounting for the origin of cells. A few highlights of each of the 12 stages will be offered next to drive this truth home, and to whet your appetite to read the full book (figure 1).
Twelve unavoidable stages for a naturalist origin of cells
1. Formation and concentration of building blocks (chapter 7)

Those interested in reviewing prebiotic chemistry research may wish to also read the classic work by a team of creation scientists, The Mystery of Life’s Origin.2
Amino acids. Unsurprising to an organic chemist, amino acids can form in small amounts under high-energy conditions using combinations of simpler molecules. The relevance to putative environments over four billion years ago is speculative, of course. The key issue is that other substances are also formed in vastly greater amounts. These can react with multiple functional groups of the amino acids, both preventing polymerization and damaging any polypeptide formed.
Phospholipids. Phospholipids are the raw material of cell membranes. Despite much effort, no one has succeeded in coming up with a construed environment with a single set of conditions able to bring together and produce large amounts of pure phosphates, glycerol, and appropriate long fatty acid chains. Remarkably, bacteria and archaea use very different phospholipids, and thus have different enzymes to produce them. This is another of many facts arguing against all life having a common ancestor.
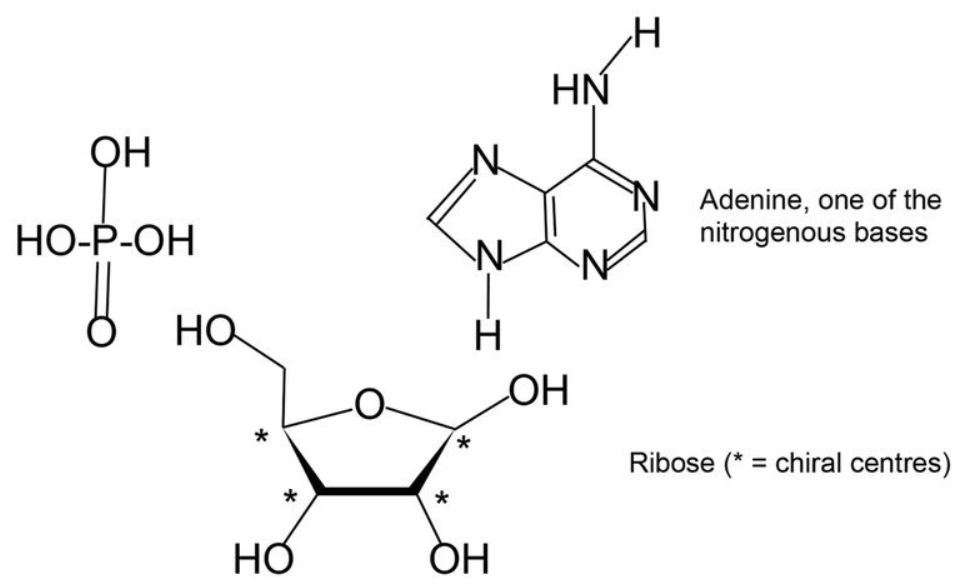
Nucleotides. Nucleotides consist of three parts: a nitrogenous base (A, C, G, or T), attached to a ribose sugar ring, and a phosphate group (figure 2). D-ribose exists in five isomeric forms (and two of those are more stable than the β-D-ribofuranose form used by DNA and RNA). Let us consider what happens when nucleotides polymerize, using adenine as a specific example. Any of the four -OH groups could react, and thus a phosphate group could react with any of the remaining three -OH groups. In addition, Adenine has three -NH locations which could bond to ribose (p. 79).
This leads to 5 × 4 × 3 × 3 = 180 possible isomeric variants per adenine nucleotide, but only one is biologically correct (p. 80). Connecting 40 nucleotides like adenine into a chain would produce 18040 = 1.6×1090 alternatives, of which only one would be biologically correct. Recall that the smallest genomes of free-living organisms have over half a million nucleotides, not 40!
Tan and Stadler take into account chirality and the need for correct nucleotide sequences later, but other experts have included these aspects also in their own calculations, cementing how absurd it is to expect natural processes to produce DNA (and RNA):
3 sites on adenine
× 5 sites on D-ribose
× 8 pentoses
× 3 sites left on pentose for phosphate links
× 2 sites left on pentose for dinucleotide links
× 3 considering that purines can form with the -NH2 at C2, C8 as well as at the C6 positions for adenine.
The number of possible dinucleotides produced naturally is 2,160.3 The true situation for evolutionists is therefore considerably worse than the calculation above showing 1.6×1090 alternatives for short chains of 40 adenines.
Given that a hypothetical RNA world is believed by many evolutionists to have preceded extant DNA-based life, much research has focused on forming ribose by polymerizing formaldehyde (the formose reaction). The desired ribose product, however, would be a minor component in a rich mixture of sugars and degrades rapidly compared to most of the other monosaccharides formed (p. 80).
All molecules needed to synthesize biomolecules must be concentrated at a suitable location but given the mishmash of co-materials present would then end up producing a tar-like mixture. When we chemists react several organic substances together at a high temperature and do not carefully separate out the products and remaining reactants, we inevitably get a blackish amorphous sticky mess we call ‘gunk’. (Which is often also foul smelling. No wonder so many otherwise normal students hate chemistry.) It takes careful planning, considerable expert knowhow, practice, and long-term controlled conditions to isolate the relevant pure molecules needed.
Many fundamental raw materials unavailable. An issue not addressed by these authors which should be kept in mind is that under various primordial scenarios the essential simpler molecules needed will be destroyed. The abiogenesis stories told are along the lines of, ‘Let’s suppose everything were just so’, but the alternative unavoidable facts are neglected.
To illustrate, UV light irradiation on Earth is believed to have been intense four billion years ago. Solar light of wavelength < 2000 Å would have caused methane to polymerize through free radical reactions and fall into the ocean as cross-linked hydrocarbons, probably covering the surface with an oil slick 1–10 metres deep. This would leave very little free methane in the atmosphere for other chemical purposes such as producing amino acids. In addition, a thick, viscous film covering the oceans would trap other organic materials, preventing the kinds of reactions needed from occurring.
About 99% of the atmospheric formaldehyde, a key ingredient in proposed routes to synthesize nucleotides and other biomolecules, would have been quickly degraded to carbon monoxide and hydrogen also by photolysis. Carbon monoxide itself would have been quickly and irreversibly converted to formate in an alkaline ocean. Any ammonia present would have been rapidly photolyzed to (very inert) nitrogen and hydrogen. Hydrogen sulfide, essential to produce some amino acids in proposed chemical scenarios, would have been photolyzed to free sulfur and hydrogen. Furthermore, any hydrogen sulfide found in an ocean would have formed insoluble metal sulfides. Photodissociation of water would have produced oxygen, and even small amounts are deadly for the chemicals used in life.4
2. Homochirality of building blocks
Many biomolecules have one or more carbon centres with the property mentioned above called chirality. The mirror images of these centres are not superimposable, like the reflection of a pair of hands. The twin molecules are called enantiomers, and based on considerations of perfect symmetry, each pair is produced in exact 50:50 proportion (with a few extremely rare exceptions), called a racemic mixture. The isomers have identical physical properties such as melting and boiling point, so none of the laboratory nor natural ways to separate them work. Separation to obtain an “optically pure” single isomer sample is done by interactions with pre-existing biomolecules from cells, which come in only one of the enantiomeric forms.
Could events with equiprobable outcomes still show predominance of one or the other? From a fundamental principle of statistics, the Law of Big Numbers, the greater the number of trials the less likely this is to occur. And the number of biomolecules needed is vast. Furthermore, in the case of biochemical enantiomers, a bond at the chiral site can temporarily break, leading to a flat intermediate to which re-bonding can occur from either side. Bond-breaking and reforming to the alternative mirror image would occur more often for the predominant variant, relentlessly driving to a perfect mixture. In fact, measuring the loss of optical purity of L-amino acids is a means of dating how long ago an organism died.
Should an unknown principle be able to form one enantiomer preferentially, the more time transpires the more opportunities to equilibrate to a perfect racemic mixture. And the increasing complexity assigned to evolution would require even more and larger optically pure building materials. Time is not the secret weapon of evolution!
The deoxyribose rings of DNA contain three chiral centres, and the RNA version has four, as shown in the figure above. The central carbon of amino acids has one such centre, and glycerol once integrated into phospholipids also have a chiral centre. This means that under natural conditions, reactions of simpler molecules leading to a chiral centre in the larger product molecule will generate a non-superimposable pair for every chiral carbon. But in living systems only one of the enantiomers must be selected to form proteins, DNA, RNA, and cell membranes, so that the final polymers can possess consistent three-dimensional structures.
The homochirality requirements are never met under natural conditions. This works in cells because the pure enantiomers are synthesized using pre-existing optically pure enzymes. And these come from, well, you know, pre-existing … . In addition, cells recycle most biomolecules continually, helping to eliminate any incorrect isomerization occurring after they were produced.
Although the need for optically pure biopolymer chains ranging from tens to millions of monomers long is well known, this unavoidable requirement is carefully overlooked in virtually all discussions of naturalistic abiogenesis.
Various attempts to circumvent the chirality problem are discussed in the book. In one case chemists artificially begin with a prevalence of one enantiomer and set up a construed autocatalytic reaction based on clever dialkylzinc chemistry having no chance of arising on its own (p. 92). Some mineral surfaces like quartz and calcite can have a minor preference for an enantiomer version of amino acids, up to 10% over a microscopic portion of the surface if the minerals are scrupulously purified (p. 92). In all these attempts one must recall that even 99% purity would not be sufficient, and most importantly that racemizing to a 50:50 mixture just occurs over time anyway.
3. A solution for the water paradox
The polymerization reactions to produce DNA, RNA, proteins, and phospholipids generate a molecule of water per chemical bond formed. However, when dissolved in water, the reaction goes in the reverse direction, to generate the individual component monomers. Water will also deaminate the nucleobases cytosine and adenine, and lead to other undesirable reactions (p. 97). In view of these established realities it is puzzling why the notion of life having arisen in a primordial ocean has not been removed from science textbooks.
Nevertheless, a suitable solvent must be available if the monomers are to react outside of cells, along with a high local energy source. Given the contradictory requirements, naturalist scenarios require clever planning by chemists who separate and protect each component to prevent destructive pathways, bring the parts together in a judicious manner, and separate the products in just the right way. An example is the use of ferricyanide, hydrogen sulfide and pure, homochiral, activated amino acids (p. 96).
4. Consistent linkage of building blocks
Key biopolymers in life are huge. On average a protein consists of about 300 amino acid monomers, and the simplest free-living organism’s DNA contains about half a million nucleotides (p. 103). Forming long ‘homo-linkage’ chains does not occur in free nature for fundamental chemical and statistical reasons. Suppose the above requirements were met, and the correct enantiomers were produced and isolated in high concentration in one location without contamination from other competing chemicals. Alas, different functional groups on each monomer (amino acid or nucleotide) can also react together, whereas only one is correct. This leads to a complex mixture known as the ‘asphalt paradox’ (p. 103).
The large biopolymers are viscous and very sticky. Long before attaining a relevant length under natural conditions they become hopelessly entangled, undergoing further internal reactions between nearby functional groups instead of only linear extensions by reacting at their ends.
5. Biopolymer reproduction
No biological life can exist without reproduction. But how are copies of DNA and RNA to be generated at all, far less reliably over millions of generations? Cells contain a collection of molecular machines to manufacture DNA, RNA, proteins, and phospholipids, and these machines consist of mainly proteins encoded on DNA. None of these manufacturing systems would be available prebiotically. Various attempts to resolve this have been carried out, but none limit themselves to components of non-biological origin. All attempts reflect a deep understanding of what sub-goals need to be met to achieve a target outcome, reflecting very clever organization and guidance. All results to date deepen our conviction that a super intelligence (God) designed cells from the very beginning.
6. Nucleotide sequences forming useful code
Even if it were possible to produce long polymers reliably this is far removed from anything with life-like features. Humans synthesize thousands of tons of plastics (which are also polymers) every year, but nobody confuses these with living organisms. Whether considering DNA as an information carrier or RNA as a source of catalysts for chemical reactions, relevant sequences of monomers must be linked together. The origin of information is a serious objection to mindless abiogenesis.
Proteins are key players, and all forms of life depend on them. Something would have had to guide the consistent reproduction of many copies of different groups of sequences. It is known that most random sequence amino acid chains serve no biological use.
7. Means of gene regulation
If all preceding steps were achieved, hundreds of RNA or DNA ‘protogenes’ would now exist, contained either in one long polymer or split up among hundreds of shorter but still very long ones. Having supposedly arisen independently via natural selection these would compete, with those having shorter reproductive cycles out-populating the others. But instead of producing only one or a few victors, the genes or living organisms must collaborate as a holistic entity.
Collections of genes must be regulated to avoid run-away production. The right number must be active at the right time and location. As far as scientists have determined, all processes in cells are regulated and in very precise ways. These involve three parts (p. 128): 1) A means of sensing; 2) a means of making decisions which are in the collective interest; and 3) a means of acting upon the decision.
Thousands of examples of feedback regulation are known, and the authors describe a few. E. coli manufactures a necessary amino acid called tryptophan using the enzyme tryptophan synthase. Producing this tryptophan consumes valuable material and energy, and runaway over-production would be deleterious. How is regulation accomplished? In the cell freely available tryptophan is involved in a negative feedback loop with tryptophan synthase. In this case, another gene is present which codes for a regulatory protein called ‘TrpR’, which can directly sense the presence of tryptophan, changing its shape (p. 129). The modified TrpR binds to a specific regulatory DNA sequence a short distance from the tryptophan synthase gene, deactivating it. Tryptophan is no longer manufactured.
The complete story was not elaborated on, but the general principle for this kind of regulation is that the regulatory proteins eventually disassociate from the cognate section of DNA. If the concentration of this protein is high, another one binds shortly afterwards, and the gene remains correctly inactivated. Otherwise the gene is unrepressed, causing more of the enzyme to be produced and thereby more of the protein identified as being in undersupply.
We notice here again the chicken-and-egg dilemma found whenever we examine regulatory schemes. What came first, the biomolecular feature or its regulatory infrastructure? This is a guiding principle I kept noticing when reading standard university textbooks on cell biology and biochemistry. It should therefore come as no surprise that non-evolutionists have no misgivings about reading any of the leading works on epigenetics,5 systems biology, 6 and biological circuits1 even if written by evolutionists. The occasional lame allegations about the wondrous guidance provided by evolution always lack plausibility or any kind of evidence, add no insight to the discussion, and are best simply ignored as irrelevant wordiness.
Until a new biological process is working close to flawlessly it would have negative value, costing valuable energy and resources, and would slow down the rate of reproduction since more DNA must be duplicated. The cellular economic tradeoffs7 of carrying incompletely developed features can be analyzed mathematically.8
8. Means for repairing biopolymers
The vast amount of information carried on DNA and RNA needs to be maintained over time. Tan and Stadler describe some sophisticated DNA repair mechanisms which counteract several ways DNA is destroyed during an organism’s lifetime in order to ensure its long-term viability (chapter 14). As one would expect, the repair processes are also coded for on DNA. Where would all this extra DNA come from, needed to eventually code for dozens of new genes? Evolutionists face the inevitable chicken-and-egg problem once more, being unwilling to accept that cells with their subsystems were designed as integrated entities from the beginning.
There is a strange notion that deep time would allow the accumulation of innumerable chunks of information. The necessary sequences are assumed to have been shorter initially, in order to ameliorate the staggering improbabilities involved. But the reality is that the physical carriers would be corrupted both during their lifetimes and for every replication cycle.
In addition, Nobel Laureate Manfred Eigen pointed out in 1971 that a prebiotic molecule like RNA could not have been very long because of the error rate during replication (p. 139). Longer polymers would introduce more errors, leading to error catastrophe. The obvious implication is that error-correcting genes could not have been added to the genome by random processes.
9. Selectively permeable membranes
Functional membranes are necessary for cells to be viable. A continuous supply of building materials and energy must by supplied from the external environment and concurrently waste products must be removed. This careful logistical control is so demanding that about a third of known proteins function within membranes (p. 143).
Pores are specialized complexes of multiple proteins working together to allow specific kinds of molecules to traverse membranes. New, evolving pores would function inappropriately leading to ‘holes’ and thereby deadly leakage. The proteins used by pores require special chemical properties not only to recognize their target substances but also to ensure their correct spatial location. Most pores require energy to function, provided in a carefully regulated manner (ATP molecules).
Membranes and their accompanying proteins are replicated and provided as a functional entity to new daughter cells (p. 147).
10. Means of harnessing energy
Cellular processes require a large amount of energy, provided in a carefully regulated manner. Sources of energy like direct blasts of UV light or high temperatures would destroy the key biochemicals. The process for harnessing and storing energy in organisms is known as ‘chemiosmotic coupling’ (p. 150). Raw energy sources include sunlight, larger organic molecules, and some inorganic simple chemicals.

The Electron Transport Chain (figure 3) is how energy is extracted from food to produce a proton gradient across a membrane (except for methanogens and acetogens, which use different, unrelated mechanisms). A sequence of about 15 reactions are linked sequentially to strip energy from carbon-containing biomolecules. This involves three protein-based complexes (NADH-Q Oxidoreductase, coenzyme Q-cytochrome c oxidoreductase, and cytochrome c oxidase). Between 25 and 69 proteins are needed, depending on the kind of organism (p. 155). Working together with an efficiency unmatched by human technology, each pair of electrons provides the energy necessary to pump 10 protons across the membrane.
On the other side of the membrane lies a generator called ATP Synthase. An excellent animation can be viewed on YouTube.9 The simplest kind known (in E. coli) requires at least 20 interconnected proteins (p. 155). This machine consumes 10 protons per three molecules of high-energy ATP molecules formed from adenosine diphosphate (ADP). ATP can then attach to carefully prepared parts of biomolecules to provide a standardized energy packet through hydrolyzation back to ADP.
Virtually everything occurring in a cell besides random diffusion requires ATP. Examples include DNA replication, disentangling DNA, charging tRNAs, translation of the genetic code, and cell replication.
11. Interdependency of DNA, RNA, and proteins
Cells require mutual collaboration between three classes of biochemicals: DNA, RNA, and proteins. The protein sequences are specified by mRNAs, which themselves are derived from DNA. Specification is not enough; the rules must be implemented, and more than 48 proteins are needed by ribosomes to carry out the translation process.
However, DNA and RNA are created from nucleotides themselves catalyzed by protein-based enzymes. Transcription of DNA to RNA requires RNA polymerases, which are mostly protein complexes, and replication of DNA requires DNA polymerases, which are also protein-based complexes of at least 14 enzymes (p. 163). There are many more examples of necessary processes DNA and RNA undergo which depend on proteins. Clearly the equipment based on DNA, RNA, and proteins must be present concurrently for a cell to be viable.
The attempt to simplify led to the poorly thought-out RNA world hypothesis. A single intractable problem was replaced by two intractable problems. Nothing has been gained for the evolutionist. The first problem involves producing an RNA form of life, very problematic given the poverty of chemical features these could offer compared to proteins.
The second problem involves transforming an RNA form of life to what we see today. A valuable insight is that there is no physical template-like relationship between RNA and functional equivalent protein sequences. The genetic code, a purely logical construct, maps the two kinds of sequences. If RNA complexes had been able to perform the tasks executed by current proteins, there is no path to convert one into the other. The extant system, based on no relationship with a putative ancestral invention would still need to be created, with all the mutually necessary subsystems using RNA, DNA, proteins, and phospholipids. Additional new requirements include needing a reliable source of ATP immediately, protein-based pores, proteins to compact DNA (p. 169), the protein-based machinery to duplicate the new cells, and so on.
12. Coordinated cellular purpose
A cell is far more than a vast organic molecular soup. If we sterilize a cell and then observe what happens, it does not suddenly act life-like (p. 171). But when a daughter cell is produced with the same components interacting correctly from the very beginning, it can then continue living on its own. Therefore no competent research team would seriously consider creating a living cell by synthesizing various molecules in the right proportions and then simply mixing them together.
Cells consist of layers of organization, with coordinated activity and crosstalk between processes in an orchestrated manner. Cells are not static and can react dynamically to external signals and current needs. But this adaptive behaviour is carefully regulated to prevent chaos. It is necessary for a multitude of individual processes to execute in a repeatable and sustainable manner at the same time from the very start.
The authors had the goal to show how:
“… every atheist, by definition, must believe that purely natural processes could surmount every step on the Stairway to Life … . One purpose of this book is to call out and clarify the leap of faith that atheism implies” (p. 185).
At each of these 12 steps an objective scientist should realize that the outcome is not what we expect to occur from chemistry nor physics acting with no intelligent input. This book belongs on the bookshelf, computer, or smartphone of all serious evolutionists and non-evolutionists.
References and notes
- The surrogate cell already had countless processes working with extreme precision. For examples of what this entails, see the anthology by Kirkwood, Rosenberger, and Galas (Eds.), Accuracy in Molecular Processes, Chapman and Hall, 1986. Return to text.
- Thaxton, C.B., Bradley, W.L., and Olsen, R.L., The Mystery of Life’s Origin: Reassessing current theories, Lewis and Stanley, Dallas, TX, 2nd edn, 1992. A newer version was released in 2020. Return to text.
- Thaxton, Bradley, and Olsen, ref. 2, p. 59. Return to text.
- Thaxton, Bradley, and Olsen, ref. 2, chap. 4. Return to text.
- Trygve Tollfsbol (Ed.), Handbook of Epigenetics: The new molecular and medical genetics, Academic Press, San Diego, CA, 2011. Return to text.
- Wallhout, M., Vidal, M., and Dekker, J. (Eds.), Handbook of Systems Biology: Concepts and insights, Academic Press, Cambridge, MA, 2013. Return to text.
- Alon, U., An Introduction to Systems Biology: Design principles of biological circuits, CRC Press, Boca Raton, FL, 2nd edn, 2020. Return to text.
- Alon, ref. 7. See part 3: Optimality. Return to text.
- youtube.com/watch?v=b_cp8MsnZFA Return to text.









Readers’ comments
Comments are automatically closed 14 days after publication.